



Next: Acknowledgements
Up: Atomistic Modeling of Biomolecular
Previous: Tools Used
Contents
We were able to calculate the complimentary error function, Cardinal B Splines, a charge mesh, the self-interaction and real space Ewald terms, and, using FFTW the library, compute the reciprocal space term. We have combined all these functions into an overreaching Particle Mesh Ewald Code that can be used to compute long-range interactions for a molecular dynamics simulation. The overall code adds the real and reciprocal space terms and subtracts the self-interaction term, yeilding the Coulombic potential of the lattice. We are currently working with a lattice of 32 atoms with varying charges, but our code is equally capable of addressing the thousands of atoms needed for large biomolecules and their systems. Because our algorithm is
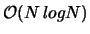 overall, it will run significantly faster for large numbers of particles than the conventional Ewald code our mentor is currently using. This should improve the feasibility of simulating biological systems with very large numbers of particles. The logical next step would be to combine our code with a subroutine that calculates short-range quantum mechanical interactions to run a molecular dynamics simulation.
overall, it will run significantly faster for large numbers of particles than the conventional Ewald code our mentor is currently using. This should improve the feasibility of simulating biological systems with very large numbers of particles. The logical next step would be to combine our code with a subroutine that calculates short-range quantum mechanical interactions to run a molecular dynamics simulation.




Next: Acknowledgements
Up: Atomistic Modeling of Biomolecular
Previous: Tools Used
Contents
Thomas G Dimiduk
2004-04-15